Defining connected fault systems
This notebook demonstrates how to identify which fault segments in a shapefile should be connected up into large fault systems.
We’ll begin by importing relevant modules
# Import modules
from fault_mesh.faults.leapfrog import LeapfrogMultiFault
import os
import numpy as np
import geopandas as gpd
Setting useful parameters
There are a few parameters that are worth setting here:
Coordinate system via EPSG code (2193 for New Zealand) — this is optional but useful if you wish to visualize contours in GIS software.
Trimming gradient (\(\alpha_{trim}\)) for clipping depth contours — see Section 3.3 of Howell et al. (in review)
Dip multiplier and strike multiplier for calculating \(\Theta_{change}\) — see Section 3.3 of Howell et al. (in review)
# Set coordinate system (optional) EPSG code
# If not desired, set to None
epsg = 2193
# Trimming gradient (alpha) and strike and dip multipliers for trimming depth contours of multi-segment faults
trimming_gradient = 1.
dip_multiplier = 1.
strike_multiplier = 0.5
Reading in faults
First, you need to read in your GIS representation of faults. In this example, we use a subset of faults from the New Zealand Community Fault Model (Seebeck et al., 2022).
# Read in fault data from shapefile
fault_data = LeapfrogMultiFault.from_shp("tutorial_gis/central_nz_minimal_data.shp", remove_colons=True, epsg=epsg, trimming_gradient=trimming_gradient,
dip_multiplier=dip_multiplier, strike_multiplier=strike_multiplier)
Finding connections between segments
Now your data are read in, you need to set the distance tolerance. This tolerance is the minimum horizontal distance between two fault traces that is allowed to count as a connection.
dist_tolerance = 200.
fault_data.segment_distance_tolerance = dist_tolerance
The next cell uses python module networkx to find segment traces that are within the specified distance tolerance of each other.
fault_data.find_connections(verbose=False)
Found 79 connections
Found 71 connections between segment ends
It is now necessary to write out these connections for manual editing and review. The file will be written out into the same directory as this Jupyter notebook. It will have a prefix supplied by you and the suffix “_suggested.csv”.
fault_data.suggest_fault_systems("central_gt1_5_connected")
This will create a CSV file that looks like this:
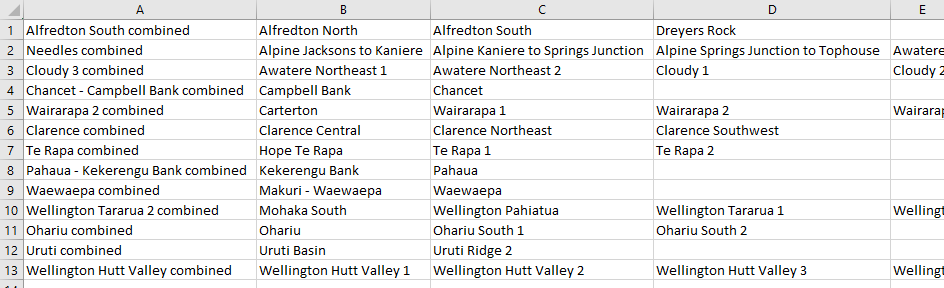
The name of the combined fault system is in the first column, and names of the faults that make up the connected system are in subsequent columns.
Making and incorporating manual edits
The automatically-generated fault system suggestions will (by design) include hyper-connected fault systems that need to be broken up. At this stage, the best way to break up these networks into smaller fault systems is to do it manually by editing the CSV file. An example below shows a new line added to the CSV representing the Hope Fault system – The Hope Fault is grouped with the Alpine and Kekerengu-Needles fault systems in the automatically-generated connections CSV. Make sure you save this new file with a different name to avoid overwriting it!
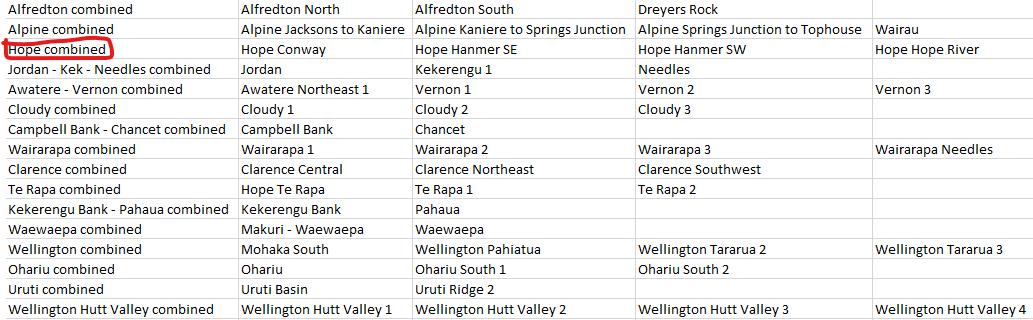
Once you have made the necessary edits, read your new CSV to overwite the automatically-generated connected fault systems:
fault_data.read_fault_systems("./define_connections_data/central_gt1_5_connected_edited.csv")
fault_data.generate_curated_faults()
Creating connected fault system: Alfredton combined with segments: ['Alfredton North', 'Alfredton South', 'Dreyers Rock']
Creating connected fault system: Alpine combined with segments: ['Alpine Jacksons to Kaniere', 'Alpine Kaniere to Springs Junction', 'Alpine Springs Junction to Tophouse', 'Wairau', 'Wairau 2', 'Wairau 3']
Creating connected fault system: Hope combined with segments: ['Hope Conway', 'Hope Hanmer SE', 'Hope Hanmer SW', 'Hope Hope River', 'Hope Hurunui', 'Hope Kakapo-2-Hamner', 'Hope Seaward', 'Kelly']
Creating connected fault system: Jordan - Kek - Needles combined with segments: ['Jordan', 'Kekerengu 1', 'Needles']
Creating connected fault system: Awatere - Vernon combined with segments: ['Awatere Northeast 1', 'Vernon 1', 'Vernon 2', 'Vernon 3', 'Vernon 4']
Creating connected fault system: Cloudy combined with segments: ['Cloudy 1', 'Cloudy 2', 'Cloudy 3']
Creating connected fault system: Campbell Bank - Chancet combined with segments: ['Campbell Bank', 'Chancet']
Creating connected fault system: Wairarapa combined with segments: ['Wairarapa 1', 'Wairarapa 2', 'Wairarapa 3', 'Wairarapa Needles']
Creating connected fault system: Clarence combined with segments: ['Clarence Central', 'Clarence Northeast', 'Clarence Southwest']
Creating connected fault system: Te Rapa combined with segments: ['Hope Te Rapa', 'Te Rapa 1', 'Te Rapa 2']
Creating connected fault system: Kekerengu Bank - Pahaua combined with segments: ['Kekerengu Bank', 'Pahaua']
Creating connected fault system: Waewaepa combined with segments: ['Makuri - Waewaepa', 'Waewaepa']
Creating connected fault system: Wellington combined with segments: ['Mohaka South', 'Wellington Pahiatua', 'Wellington Tararua 2', 'Wellington Tararua 3', 'Wellington Tararua 1']
Creating connected fault system: Ohariu combined with segments: ['Ohariu', 'Ohariu South 1', 'Ohariu South 2']
Creating connected fault system: Uruti combined with segments: ['Uruti Basin', 'Uruti Ridge 2']
Creating connected fault system: Wellington Hutt Valley combined with segments: ['Wellington Hutt Valley 1', 'Wellington Hutt Valley 2', 'Wellington Hutt Valley 3', 'Wellington Hutt Valley 4', 'Wellington Hutt Valley 5']
Defining a cutting hierarchy
Now you have defined your fault system (which you will later use to create meshes), it is necessary to specify which faults terminate against other faults. For example, it seems highly likely that the western end of the Hope Fault is truncated at depth by the Alpine fault. This complex mesh cutting is best achieved using dedicated software such as leapfrog (see below), but for cutting to be done automatically, it is best to first specify a cutting hierarchy.
Suggesting a hierarchy based on slip rate
A first pass at a hierarchy can be generated based purely on fault slip rate. Assuming that the fault data you have already read in have slip rates associated with them, this first pass is easy to make:
fault_data.suggest_cutting_hierarchy("central_gt1_5_hierarchy")
This operation simply orders the faults (or fault systems) in your model in descending order of slip rate. For connected fault systems that have segments with different slip rates, the maximum slip rate of any segment in that fault system is used to place the fault system in the cutting hierarchy.
Editing the cutting hierarchy
You can then edit this hierarchy by switching the order of lines in the file. For any pair of faults/systems that intersect, the fault closer to the bottom of the file will terminate against the fault closer to the top of the list.
An example of a situation where editing is desirable is illustrated below. The maximum slip rate of the Jordan-Kekerengu-Needles Fault System (23 mm/yr) is faster than the corresponding maximum for the Hope Fault (15.8 mm/yr), but we wish to create a fault model where the Jordan Fault terminates against the Hope Fault. We effect this termination by moving Hope combined above Jordan - Kek - Needles combined in the CSV file. For similar reasons, we move the Hanmer Fault above Hope Hanmer NW.
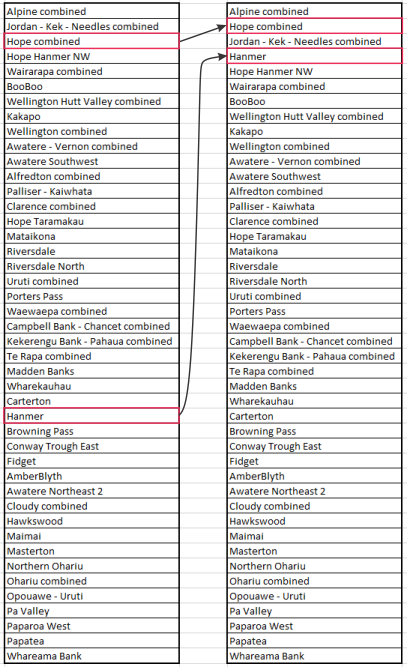
We read in this new hierarchy as follows:
fault_data.read_cutting_hierarchy("./define_connections_data/central_gt1_5_hierarchy_edited.csv")
Create shapefiles for mesh creation
The final pre-meshing step is the creation of files that can be combined with meshing software to create triangular mesh representations of faults. Although it is possible to construct these triangular surfaces in multiple software packages (for example, MOVE 3D), the following discussion is geared towards use with Leapfrog Geo software.
Create directories to hold shapefiles
For organisational reasons, it helps to have the different shapefiles in different directories
for dir_name in ["depth_contours", "traces", "footprints", "footprints_lines"]:
if not os.path.exists(dir_name):
os.mkdir(dir_name)
Write out shapefiles
for fault in fault_data.curated_faults:
# Generate depth contours
fault.generate_depth_contours(np.arange(2000, 32000., 2000.), smoothing=False)
# Write contours to files
fault.contours.to_file(f"depth_contours/{fault.name}_contours.shp")
# Write traces
fault.nztm_trace_geoseries.to_file(f"traces/{fault.name}_trace.shp")
# Write fault footprints
for fault in reversed(fault_data.curated_faults):
fault.adjust_footprint()
fault.footprint_geoseries.to_file(f"footprints/{fault.name}_footprint.shp")
# Write fault footprints as lines (necessary for newer Leapfrog Geo betas)
fault.footprint_geoseries.boundary.to_file(f"footprints_lines/{fault.name}_footprint.shp")
fault.segments[0].dip_max